Admixture Graphs
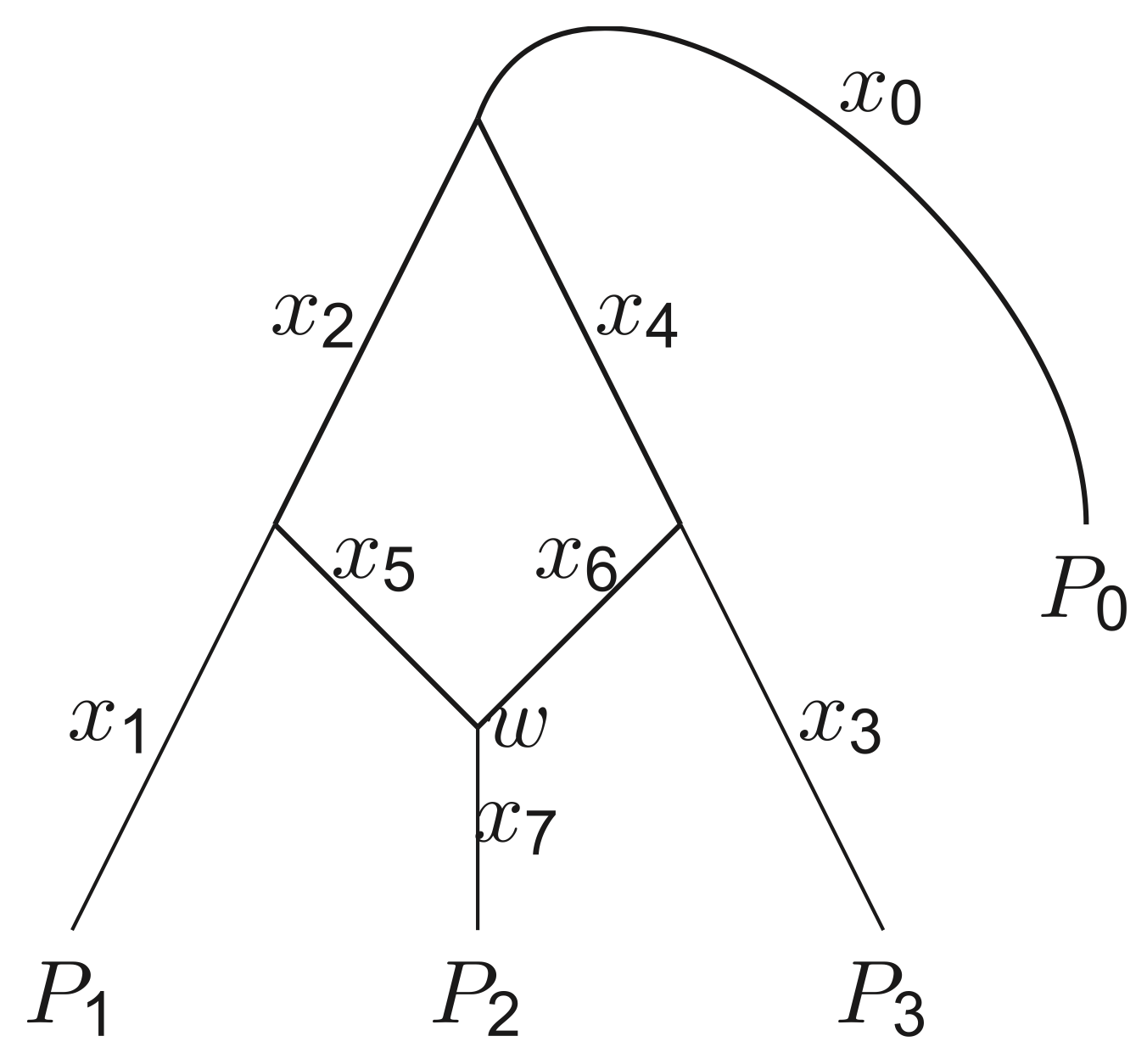
Admixture graphs provide a
concise description of the historical demographic relationships between
genetic samples of populations, assuming their relationships are the
product of discrete, instantaneous splits and admixture events. Each
admixture graph topology is associated with parameters capturing genetic drift
and admixture proportions, and once these are fitted to genetic data,
the goodness of fit can be measured to determine how accurately the
graph captures the historical relationship between samples. Inferring
graph topologies, however, involves a combinatorial search, and
since the space of graphs grows super-exponentially in the number
of populations and the number of admixture events, an exhaustive
search is typically not possible. Instead, the search for well-fitting
topologies is often done manually or through greedy algorithms, which
are not guaranteed to find a global optimum. This motivated the development of
a novel MCMC sampling method,
AdmixtureBayes,
that can sample from the posterior distribution of admixture graphs.
This enables an effective search of the entire state space as well as
the ability to report a level of confidence in the sampled graphs.
We are actively looking at ways to improve AdmixtureBayes by using
more information in the data than just the
covariance in allele frequencies between populations.
Ancestral Recombination Graphs
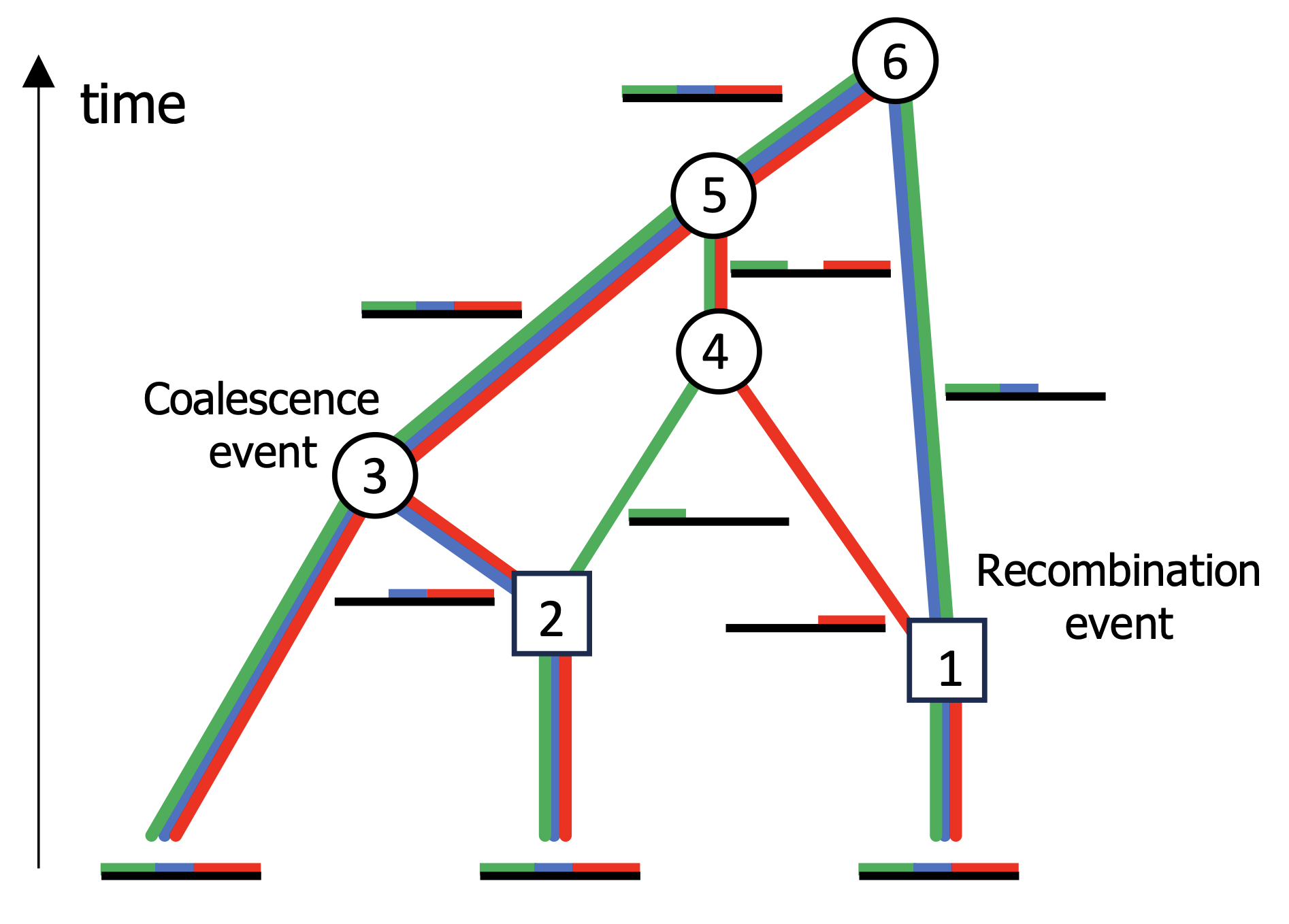
Ancestral recombination graphs (ARGs) represent the history of coalescence and
recombination events in the history of a sample of DNA sequences. There is currently
significant interest in both the development of methods to infer ARGs from sequence
data and the application of ARGs to estimate population genetic parameters. My research
has focused mostly on the latter, specifically the development of methods to infer
demographic history and natural selection from DNA using ARGs. I am the lead developer and
current maintainer of the
CLUES2 software, a method to
infer selection coefficients and historic allele frequency trajectories using inferred ARGs.
CLUES2 can infer time-varying selection coefficients, selection under arbitrary dominance
scenarios, and can work with ARGs inferred on ancient DNA. I also have an ongoing project on using
ARGs to infer patterns of human migration and dispersal.
Analyses of Ancient DNA
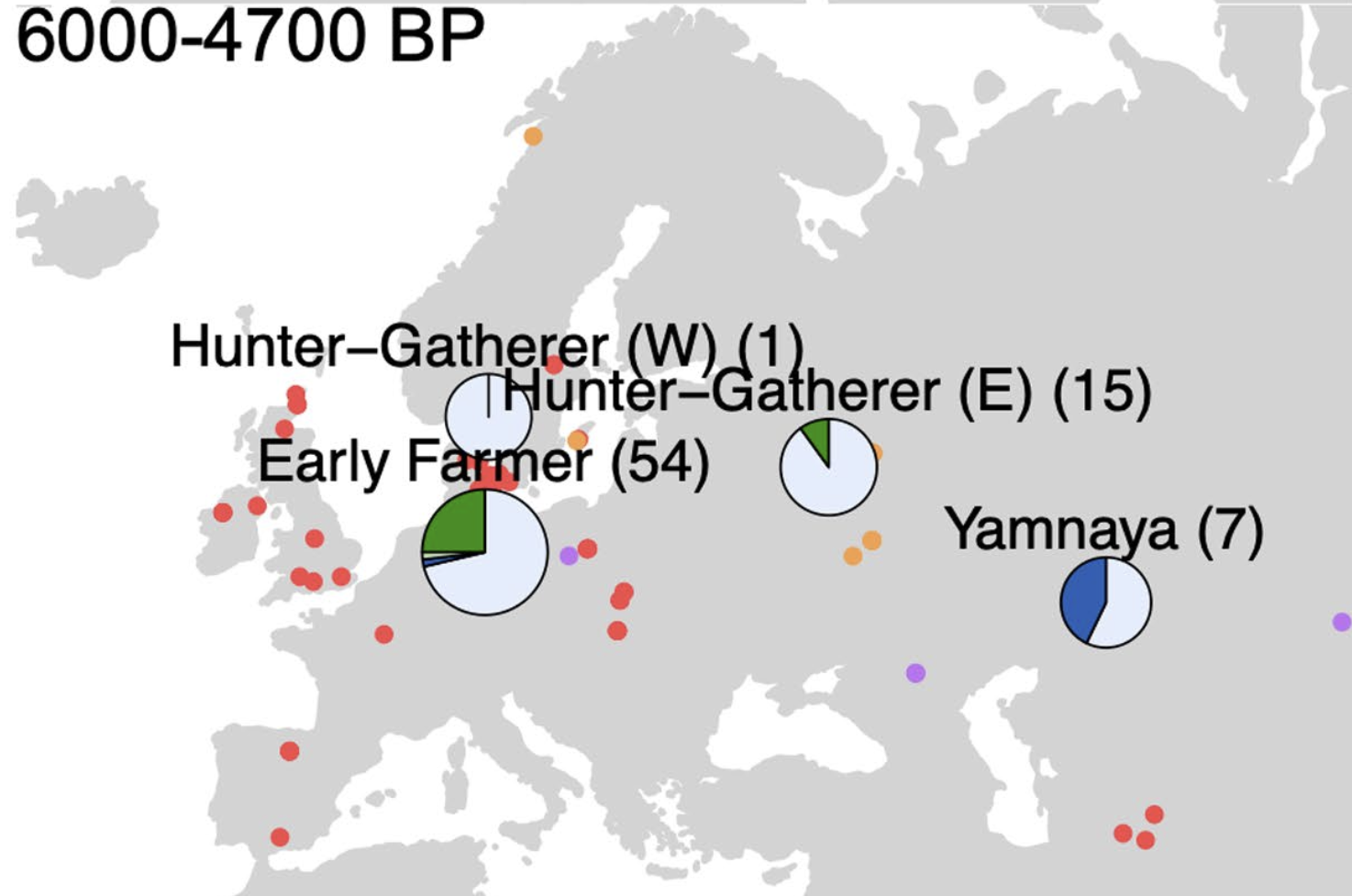
I work closely with researchers at
the
Lundbeck
Foundation Center for GeoGenetics at the University of Copenhagen
on analyzing ancient human DNA. Specifically, I have worked on studying ancestry-specific
selective pressures in Stone Age Eurasian populations and quantifying the ability
to accurately infer ARGs on low-coverage data. I have also re-analyzed several
datasets generated by the Center for GeoGenetics for both the CLUES2 and
AdmixtureBayes projects, generating new insights into changing selective pressures on
diet-associated alleles and the peopling of the Americas respectively.
Human Adaptation and Evolution
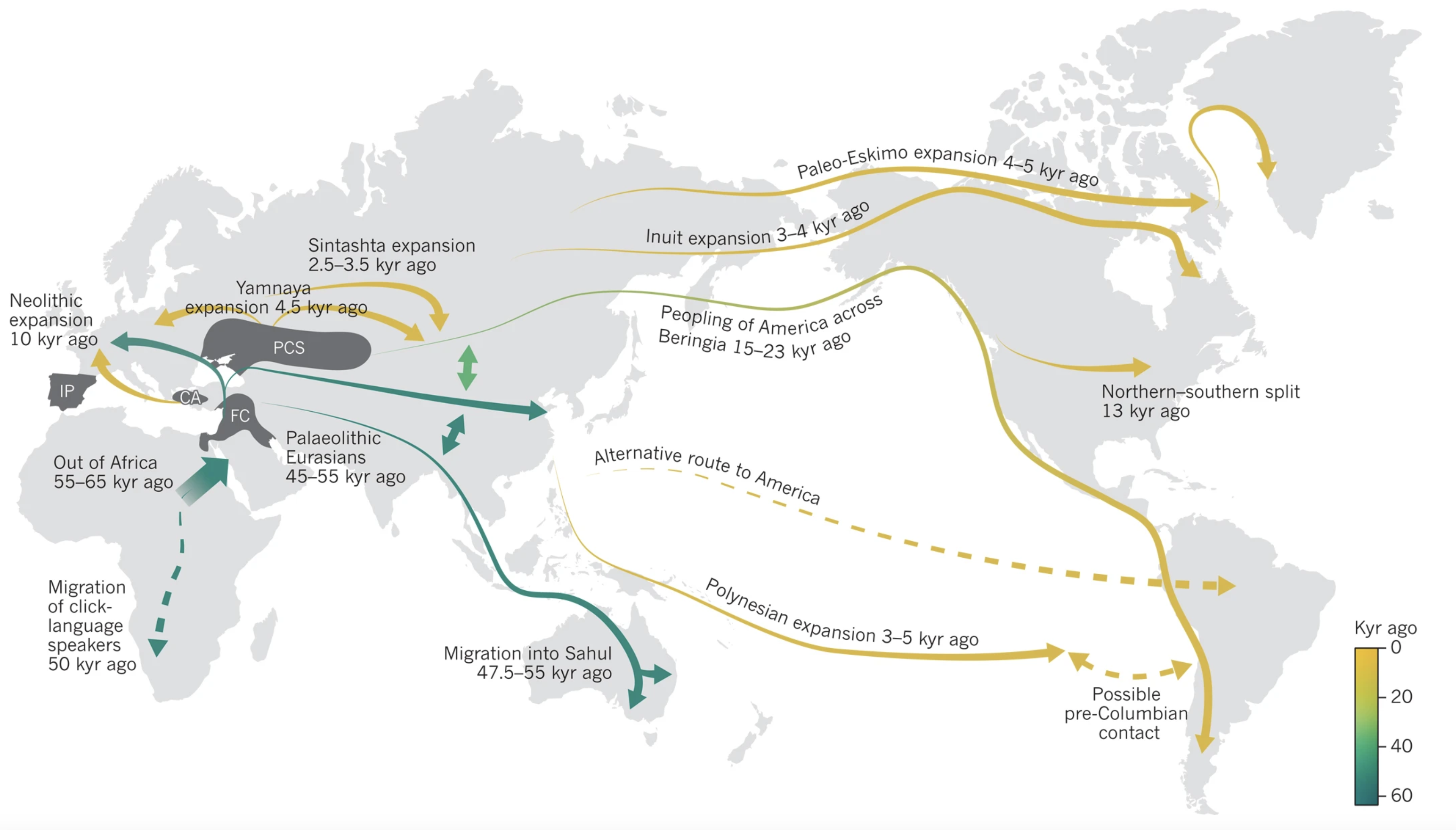
Much of the work I do on ARGs, methods development, and ancient DNA analysis is motivated by the desire
to better understand how demographic events and selective forces have shaped the
evolution of the human genome. I am particularly interested in how dietary and other lifestyle changes have affected
the evolution of metabolic traits and how these changes continue to affect human health today. My main project
in this area concerns studying the pleiotropic effect of certain alleles on
adverse health outcomes such as type 2 diabetes and coronary artery disease. Specifically,
we have found evidence that risk alleles for these diseases may have been selected for in the past, when
human lifestyles were quite distinct from those of today. Therefore, the same alleles that were
advantageous in the past may now be detrimental to health, through an evolutionary mismatch
of ancient and modern lifestyles.
 Admixture graphs provide a
concise description of the historical demographic relationships between
genetic samples of populations, assuming their relationships are the
product of discrete, instantaneous splits and admixture events. Each
admixture graph topology is associated with parameters capturing genetic drift
and admixture proportions, and once these are fitted to genetic data,
the goodness of fit can be measured to determine how accurately the
graph captures the historical relationship between samples. Inferring
graph topologies, however, involves a combinatorial search, and
since the space of graphs grows super-exponentially in the number
of populations and the number of admixture events, an exhaustive
search is typically not possible. Instead, the search for well-fitting
topologies is often done manually or through greedy algorithms, which
are not guaranteed to find a global optimum. This motivated the development of
a novel MCMC sampling method, AdmixtureBayes,
that can sample from the posterior distribution of admixture graphs.
This enables an effective search of the entire state space as well as
the ability to report a level of confidence in the sampled graphs.
We are actively looking at ways to improve AdmixtureBayes by using
more information in the data than just the
covariance in allele frequencies between populations.
Admixture graphs provide a
concise description of the historical demographic relationships between
genetic samples of populations, assuming their relationships are the
product of discrete, instantaneous splits and admixture events. Each
admixture graph topology is associated with parameters capturing genetic drift
and admixture proportions, and once these are fitted to genetic data,
the goodness of fit can be measured to determine how accurately the
graph captures the historical relationship between samples. Inferring
graph topologies, however, involves a combinatorial search, and
since the space of graphs grows super-exponentially in the number
of populations and the number of admixture events, an exhaustive
search is typically not possible. Instead, the search for well-fitting
topologies is often done manually or through greedy algorithms, which
are not guaranteed to find a global optimum. This motivated the development of
a novel MCMC sampling method, AdmixtureBayes,
that can sample from the posterior distribution of admixture graphs.
This enables an effective search of the entire state space as well as
the ability to report a level of confidence in the sampled graphs.
We are actively looking at ways to improve AdmixtureBayes by using
more information in the data than just the
covariance in allele frequencies between populations.
 Ancestral recombination graphs (ARGs) represent the history of coalescence and
recombination events in the history of a sample of DNA sequences. There is currently
significant interest in both the development of methods to infer ARGs from sequence
data and the application of ARGs to estimate population genetic parameters. My research
has focused mostly on the latter, specifically the development of methods to infer
demographic history and natural selection from DNA using ARGs. I am the lead developer and
current maintainer of the CLUES2 software, a method to
infer selection coefficients and historic allele frequency trajectories using inferred ARGs.
CLUES2 can infer time-varying selection coefficients, selection under arbitrary dominance
scenarios, and can work with ARGs inferred on ancient DNA. I also have an ongoing project on using
ARGs to infer patterns of human migration and dispersal.
Ancestral recombination graphs (ARGs) represent the history of coalescence and
recombination events in the history of a sample of DNA sequences. There is currently
significant interest in both the development of methods to infer ARGs from sequence
data and the application of ARGs to estimate population genetic parameters. My research
has focused mostly on the latter, specifically the development of methods to infer
demographic history and natural selection from DNA using ARGs. I am the lead developer and
current maintainer of the CLUES2 software, a method to
infer selection coefficients and historic allele frequency trajectories using inferred ARGs.
CLUES2 can infer time-varying selection coefficients, selection under arbitrary dominance
scenarios, and can work with ARGs inferred on ancient DNA. I also have an ongoing project on using
ARGs to infer patterns of human migration and dispersal.
 I work closely with researchers at
the Lundbeck
Foundation Center for GeoGenetics at the University of Copenhagen
on analyzing ancient human DNA. Specifically, I have worked on studying ancestry-specific
selective pressures in Stone Age Eurasian populations and quantifying the ability
to accurately infer ARGs on low-coverage data. I have also re-analyzed several
datasets generated by the Center for GeoGenetics for both the CLUES2 and
AdmixtureBayes projects, generating new insights into changing selective pressures on
diet-associated alleles and the peopling of the Americas respectively.
I work closely with researchers at
the Lundbeck
Foundation Center for GeoGenetics at the University of Copenhagen
on analyzing ancient human DNA. Specifically, I have worked on studying ancestry-specific
selective pressures in Stone Age Eurasian populations and quantifying the ability
to accurately infer ARGs on low-coverage data. I have also re-analyzed several
datasets generated by the Center for GeoGenetics for both the CLUES2 and
AdmixtureBayes projects, generating new insights into changing selective pressures on
diet-associated alleles and the peopling of the Americas respectively.
 Much of the work I do on ARGs, methods development, and ancient DNA analysis is motivated by the desire
to better understand how demographic events and selective forces have shaped the
evolution of the human genome. I am particularly interested in how dietary and other lifestyle changes have affected
the evolution of metabolic traits and how these changes continue to affect human health today. My main project
in this area concerns studying the pleiotropic effect of certain alleles on
adverse health outcomes such as type 2 diabetes and coronary artery disease. Specifically,
we have found evidence that risk alleles for these diseases may have been selected for in the past, when
human lifestyles were quite distinct from those of today. Therefore, the same alleles that were
advantageous in the past may now be detrimental to health, through an evolutionary mismatch
of ancient and modern lifestyles.
Much of the work I do on ARGs, methods development, and ancient DNA analysis is motivated by the desire
to better understand how demographic events and selective forces have shaped the
evolution of the human genome. I am particularly interested in how dietary and other lifestyle changes have affected
the evolution of metabolic traits and how these changes continue to affect human health today. My main project
in this area concerns studying the pleiotropic effect of certain alleles on
adverse health outcomes such as type 2 diabetes and coronary artery disease. Specifically,
we have found evidence that risk alleles for these diseases may have been selected for in the past, when
human lifestyles were quite distinct from those of today. Therefore, the same alleles that were
advantageous in the past may now be detrimental to health, through an evolutionary mismatch
of ancient and modern lifestyles.